# attaching packages ####
library(igraph)
library(netplot)
library(data.table)Middle School Students
This webpage will explore the dataset exploring connections in 7th and 8th grade students at a middle school. The data was taken from “Estimates of Social Contact in a Middle School Based on Self-Report and Wireless Sensor Data”. This is the “Example 1” on the poster.
Introduction || Cleaning Data || Explore Data
These are the packages that were used for this analysis:
First, let’s clean it all up, getting rid of the “isolates” (those who only are connected to one person). Here is the script for cleaning that data:
# loading and cleaning data ####
students <- fread("./misc/data-raw/middle_school/pone.0153690.s001.csv")
interactions <- fread("./misc/data-raw/middle_school/pone.0153690.s003.csv")
students <- students[!is.na(id)]
students <- students[gender %in% c("0", "1")]
interactions <- interactions[!is.na(id) & !is.na(contactId)]
## Which connections are not OK?
ids <- sort(unique(students$id))
## Alright, this narrowed our data from 10781 to 5150
interactions <- interactions[(id %in% ids) & (contactId %in% ids)]
## Creating weights matrix
net <- graph_from_data_frame(
d = interactions[, .(id, contactId)],
directed = FALSE, vertices = as.data.frame(students)
)
## Getting only connected individuals
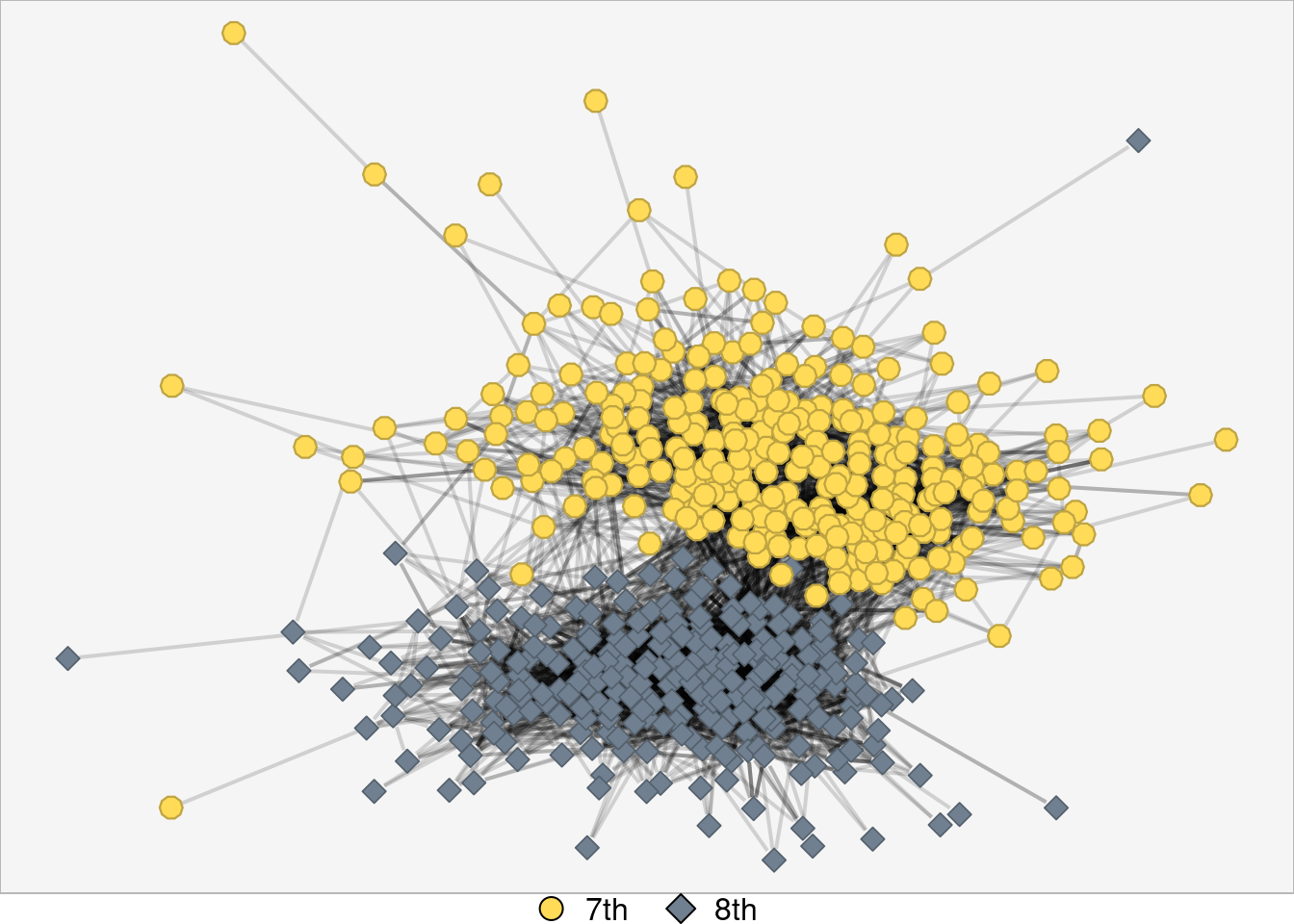
net_with_no_isolates <- induced_subgraph(net, which(degree(net) > 0))
## Plot with no isolates
nplot(
net_with_no_isolates
) 
Awesome. Now that things are cleaned up a bit, let’s see what separate sections we are dealing with!
# let's see what it looks like ####
print(net_with_no_isolates)IGRAPH 5f80021 UN-- 541 5136 --
+ attr: name (v/c), grade (v/n), gender (v/n), unique (v/n), lunch
| (v/n), initialsNum (v/n)
+ edges from 5f80021 (vertex names):
[1] 2004--3127 2004--2620 2004--2141 2004--2362 2004--2362 2004--2294
[7] 2004--2274 2004--2402 2004--2402 2004--2402 2004--2603 2004--2603
[13] 2004--2603 2004--2603 2004--2603 2004--2603 2004--2603 2004--2603
[19] 2004--2603 2004--2028 2004--2028 2004--2028 2004--2308 2004--3025
[25] 2006--2028 2006--2028 2006--2028 2006--3158 2006--3381 2006--2495
[31] 2006--2495 2006--2494 2006--2356 2006--2356 2006--2408 2009--2346
[37] 2009--3018 2009--2265 2009--2395 2009--2395 2009--3134 2009--3427
+ ... omitted several edgesHere we go! Other than the new unique aspects, we have grade, gender, and lunch period that we can explore.
Load in “color_nodes.R” function
## load in 'color_nodes' function ####
source(file = "./misc/color_nodes_function.R")Splitting Data
Split According to Grade
## adjust 'grade' to factor
V(net_with_no_isolates)$grade <- as.factor(V(net_with_no_isolates)$grade)
# plotting connections among grades ####
set.seed(77)
a_colors <- color_nodes(net_with_no_isolates,"grade", c("gray40","red3"))
attr(a_colors, "map") 7 8
"#666666" "#CD0000" grades <- nplot(
net_with_no_isolates,
vertex.color = color_nodes(net_with_no_isolates, "grade", c("gray40","red3")),
vertex.nsides = ifelse(V(net_with_no_isolates)$grade == 7, 10, 10),
vertex.size.range = c(0.015, 0.015),
edge.color = ~ego(alpha = 1, col = "lightgray") + alter(alpha = 0.25, col = "lightgray"),
vertex.label = NULL,
edge.curvature = pi/6,
edge.line.breaks = 10
)
# add radial gradient fill
grades <- set_vertex_gpar(grades,
element = "core",
fill = lapply(get_vertex_gpar(grades, "frame", "col")$col, \(i) {
radialGradient(c("white", i), cx1=.8, cy1=.8, r1=0)
}))
# add legend to graph
grades_general <- nplot_legend(
grades,
labels = c("7th", "8th"),
pch = c(21,21),
gp = gpar(
fill = c("gray40","red3")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grades_general
grid.text("Split According to Grade", x = .2, y = .87, just = "bottom") 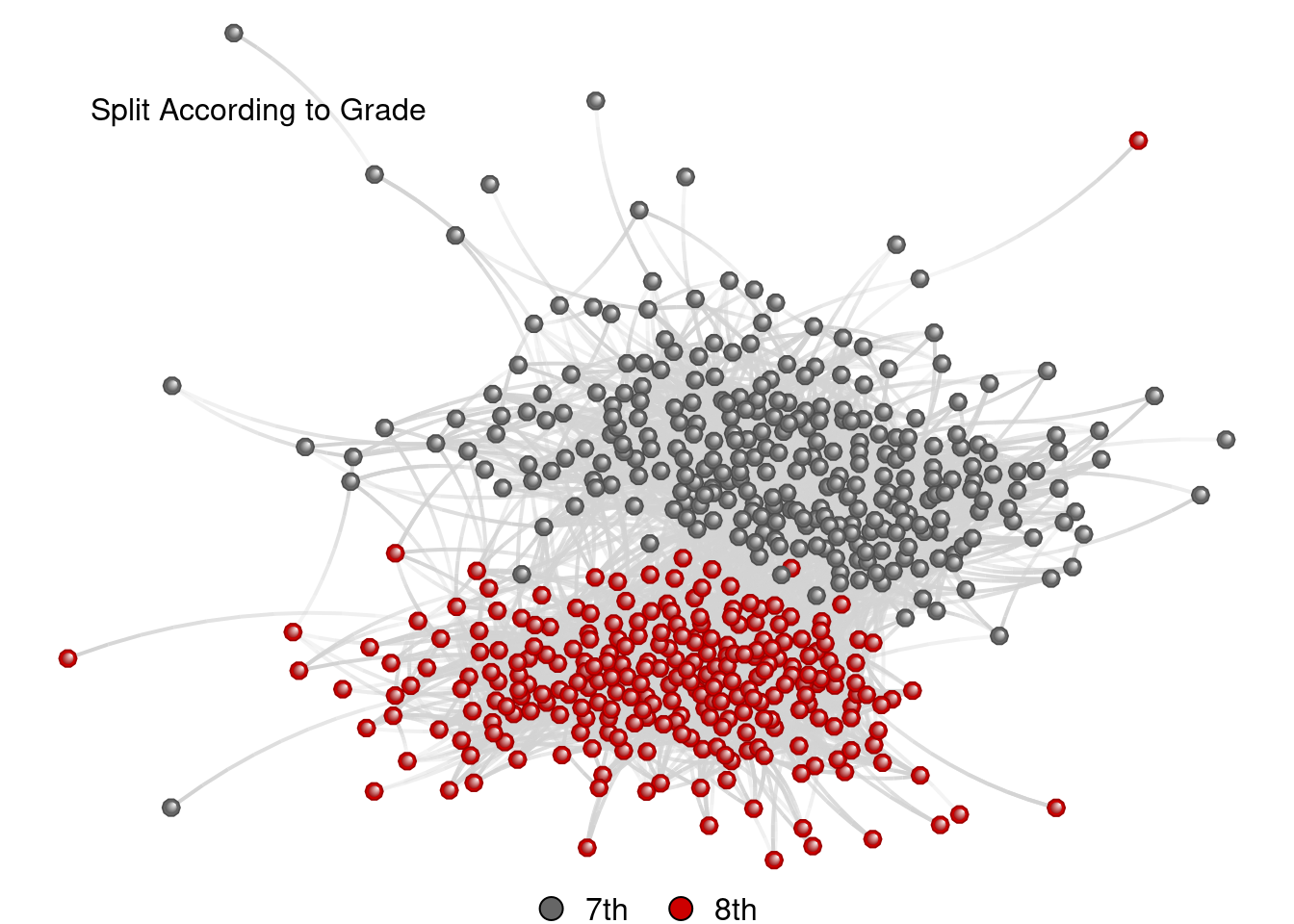
Split According to Gender
# let's get a graph for the gender data
V(net_with_no_isolates)$gender <- as.factor(V(net_with_no_isolates)$gender)
a_colors <- color_nodes(net_with_no_isolates,"gender", c("lightgoldenrod2","forestgreen"))
attr(a_colors, "map") 0 1
"#EEDC82" "#228B22" ## plot
set.seed(77)
gender <- nplot(
net_with_no_isolates,
vertex.color = color_nodes(net_with_no_isolates, "gender",c("lightgoldenrod2","forestgreen")),
vertex.nsides = ifelse(V(net_with_no_isolates)$gender == 0, 10, 4),
vertex.size.range = c(0.01, 0.01),
edge.color = ~ego(alpha = 0.33, col = "gray") + alter(alpha = 0.33, col = "gray"),
vertex.label = NULL,
edge.line.breaks = 10
)
# add legend to graph
nplot_legend(
gender,
labels = c("Male", "Female"),
pch = c(21,23),
gp = gpar(
fill = c("lightgoldenrod2","forestgreen")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grid.text("Split According to Gender", x = .2, y = .87, just = "bottom") 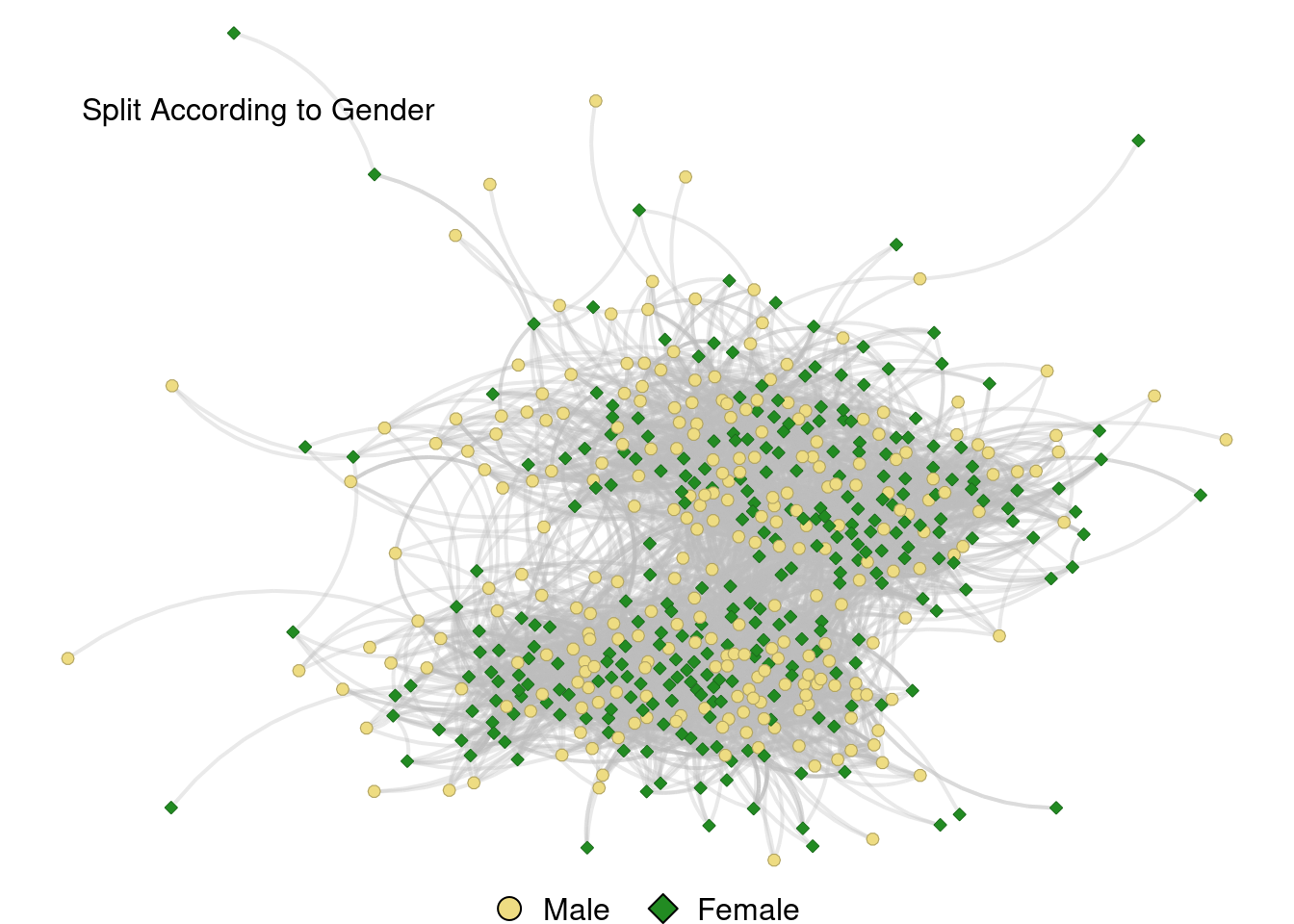
Split According to Lunch Period
# now let's do the same with lunch period
V(net_with_no_isolates)$lunch <- as.factor(V(net_with_no_isolates)$lunch)
a_colors <- color_nodes(net_with_no_isolates,"lunch", c("purple","palegreen","steelblue"))
attr(a_colors, "map") 1 2 99
"#A020F0" "#98FB98" "#4682B4" ## plot
set.seed(77)
lunch <- nplot(
net_with_no_isolates,
vertex.color = color_nodes(net_with_no_isolates, "lunch",c("purple","palegreen","steelblue")),
vertex.nsides =
ifelse(V(net_with_no_isolates)$gender == 0, 4, # First Lunch
ifelse(V(net_with_no_isolates)$gender == 1, 3, # Second Lunch
10)), # Other
vertex.size.range = c(0.01, 0.01),
edge.color = ~ego(alpha = 0.33, col = "gray") + alter(alpha = 0.33, col = "gray"),
vertex.label = NULL,
edge.line.breaks = 10
)
# add legend to graph
nplot_legend(
lunch,
labels = c("First", "Second", "Other"),
pch = c(23,24,21),
gp = gpar(
fill = c("purple","palegreen","steelblue")),
packgrob.args = list(side = "bottom"),
ncol = 3
)
grid.text("Split According to Lunch Period", x = .2, y = .87, just = "bottom") 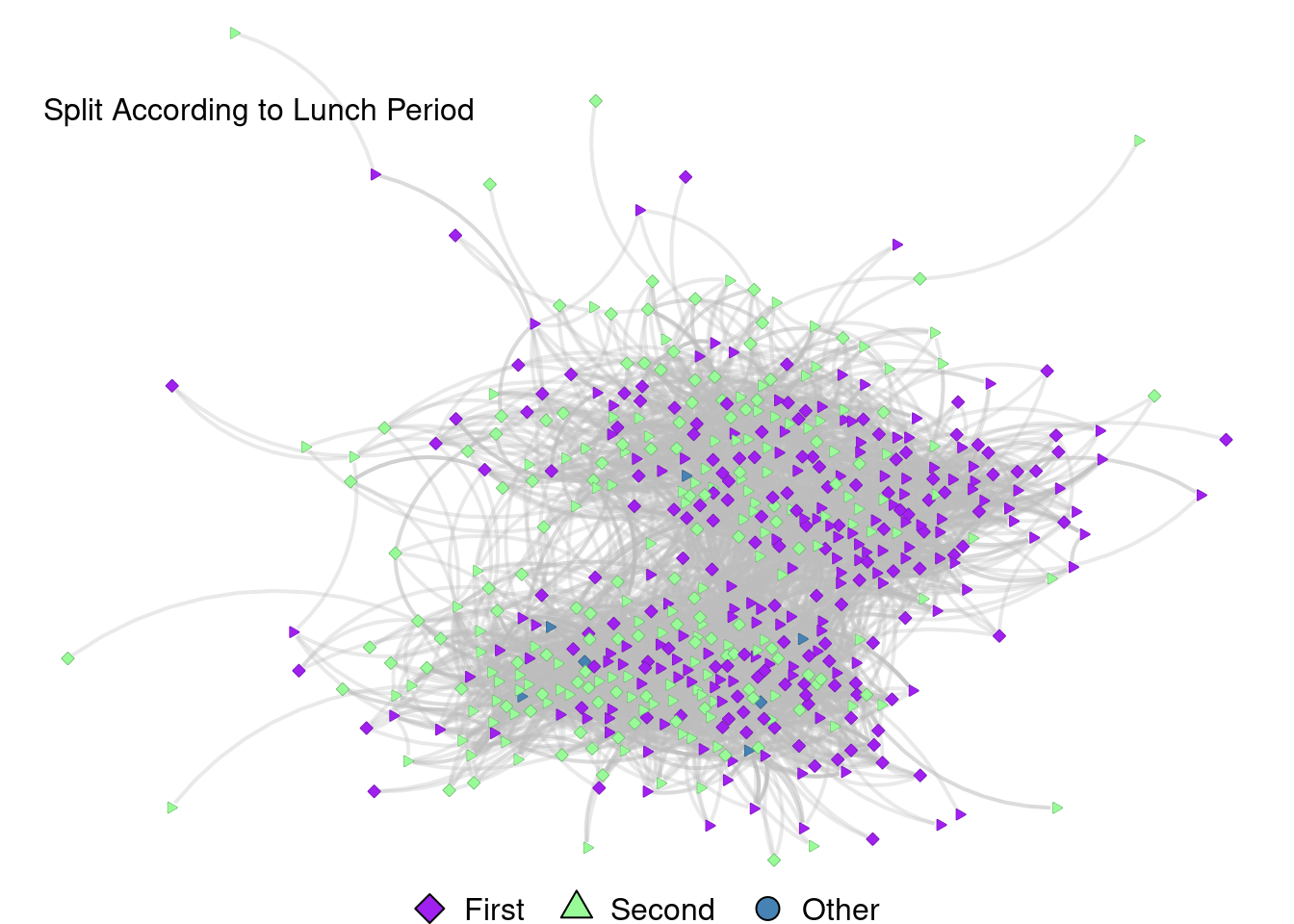
Different netplot Options
Changing Lines to Dashes
This graph correlates to the graph titled “Changing lines to dashes, diamonds to circles, and edge lines to straight lines” on the poster.
set.seed(77)
grades <- nplot(
net_with_no_isolates,
bg.col = "#F5F5F5",
vertex.color = color_nodes(net_with_no_isolates, "grade", c("red","blue")),
vertex.size.range = c(0.02, 0.02),
edge.color = ~ego(alpha = .15, col = "black") + alter(alpha = .15, col = "black"),
vertex.label = NULL,
edge.width.range = c(2,2),
edge.line.lty = 6,
edge.line.breaks = 1
)
# add legend to graph
grades_dashed <- nplot_legend(
grades,
labels = c("7th", "8th"),
pch = c(21,21),
gp = gpar(
fill = c("red","blue")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grades_dashed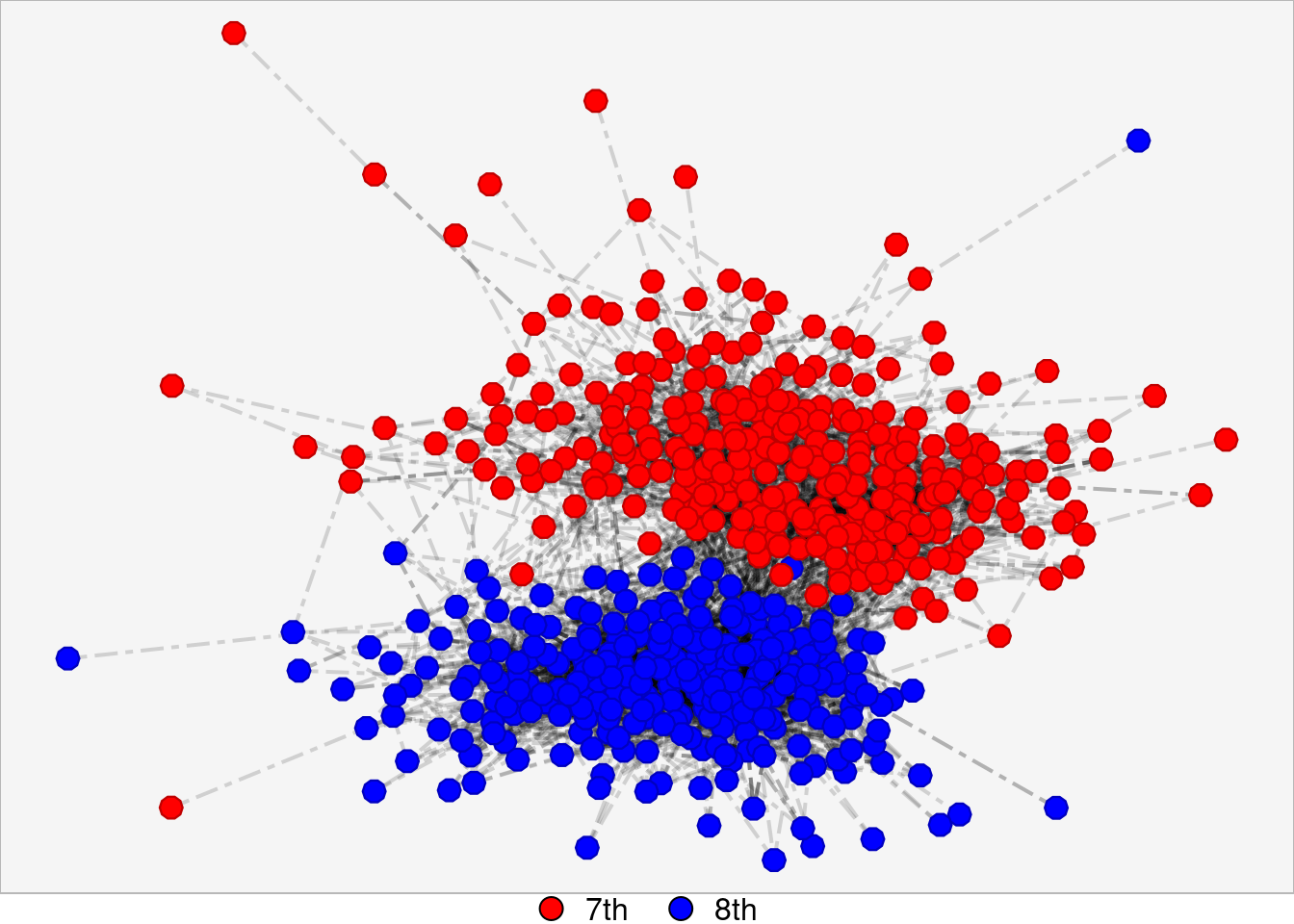
Colored Edges and Skipped Vertices
This graph is correlated to the graph titled “Skipping vertices” on the poster.
set.seed(77)
grades <- nplot(
net_with_no_isolates,
bg.col = "#F5F5F5",
vertex.color = color_nodes(net_with_no_isolates, "grade", c("red","blue")),
vertex.nsides = ifelse(V(net_with_no_isolates)$grade == 7, 10, 4),
vertex.size.range = c(0.0001, 0.0001),
edge.color = ~ego(alpha = 0.33) + alter(alpha = 0.33),
vertex.label = NULL,
edge.width.range = c(2,2),
edge.line.breaks = 10
)
# add legend to graph
grades_edge_colored <- nplot_legend(
grades,
labels = c("7th", "8th"),
pch = c(21,21),
gp = gpar(
fill = c("red","blue")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grades_edge_colored
Changing Background Color
This graph correlates with the graph titled “Background gradient as function of vertex color” on the poster.
set.seed(77)
grades <- nplot(
net_with_no_isolates,
bg.col = linearGradient(c("lightpink", "lightskyblue")),
vertex.color = color_nodes(net_with_no_isolates, "grade", c("red","blue")),
vertex.nsides = ifelse(V(net_with_no_isolates)$grade == 7, 10, 4),
vertex.size.range = c(0.01, 0.01),
edge.color = ~ego(alpha = 0.15, col = "black") + alter(alpha = 0.15, col = "black"),
vertex.label = NULL,
edge.line.breaks = 10
)
# add legend to graph
grades_background <- nplot_legend(
grades,
labels = c("7th", "8th"),
pch = c(21,23),
gp = gpar(
fill = c("red","blue")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grades_background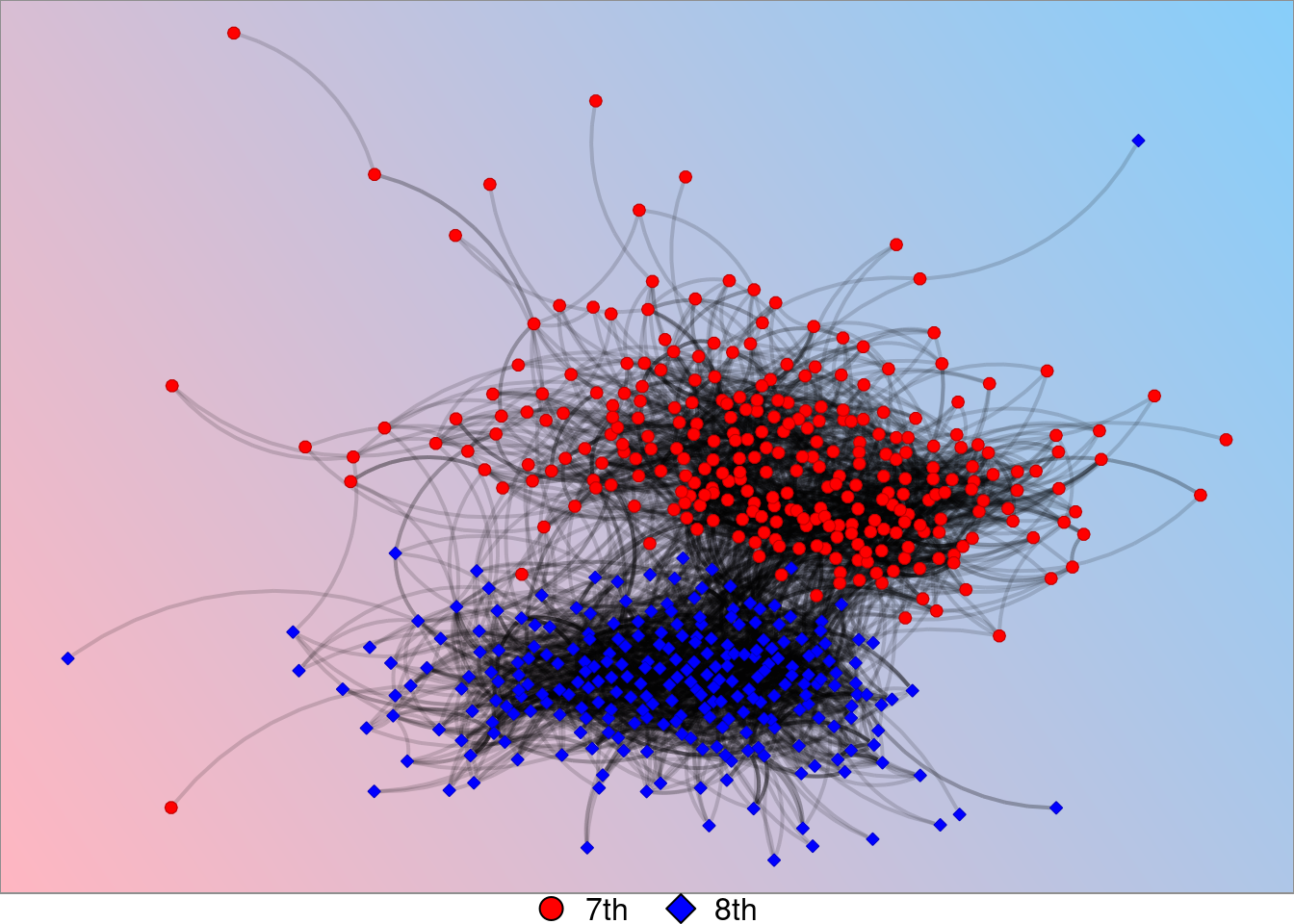
Different Colors
This graph correlates to the graph titled “Changing vertex colors & edge lines to straight lines” on the poster.
set.seed(77)
grades <- nplot(
net_with_no_isolates,
bg.col = "#F5F5F5",
vertex.color = color_nodes(net_with_no_isolates, "grade", c("#FFDB58","#708090")),
vertex.nsides = ifelse(V(net_with_no_isolates)$grade == 7, 10, 4),
vertex.size.range = c(0.02, 0.02),
edge.color = ~ego(alpha = .15, col = "black") + alter(alpha = .15, col = "black"),
vertex.label = NULL,
edge.width.range = c(2,2),
edge.line.breaks = 1
)
# add legend to graph
grades_different_color <- nplot_legend(
grades,
labels = c("7th", "8th"),
pch = c(21,23),
gp = gpar(
fill = c("#FFDB58","#708090")),
packgrob.args = list(side = "bottom"),
ncol = 2
)
grades_different_color