
Run Multiple
Derek Meyer
George Vega Yon
2026-02-18
Source:vignettes/run-multiple.Rmd
run-multiple.RmdIntroduction
The purpose of the “run_multiple” function is to run a specified number of simulations using the same model object. That is, this function makes it possible to compare model results across several separate and repeated simulations.
Example: Simulating a SEIRCONN Model 50 Times
Setup and Running Model
To use the “run_multiple” function in epiworld, create your epimodel of choice; in this case, the example uses a SEIRCONN model for COVID-19, 100000 people, an initial prevalence of 0.0001 (0.01%), a contact rate of 2, probability of transmission 0.5, a total of 7 incubation days, and probability of recovery .
library(epiworldR)
#> Thank you for using epiworldR! Please consider citing it in your work.
#> You can find the citation information by running
#> citation("epiworldR")
model_seirconn <- ModelSEIRCONN(
name = "COVID-19",
n = 10000,
prevalence = 0.0001,
contact_rate = 2,
transmission_rate = 0.5,
incubation_days = 7,
recovery_rate = 1 / 3
)Generating a Saver
Next, generate a saver for the purpose of extracting the “total_hist” and “reproductive” information from the model object. Now, use the “run_multiple” function with the model object, number of desired days to run the simulation, number of simulations to run, and number of threads for parallel computing.
# Generating a saver
saver <- make_saver("total_hist", "reproductive")
# Running and printing
run_multiple(model_seirconn, ndays = 50, nsims = 50, saver = saver, nthreads = 2)
#> Starting multiple runs (50) using 2 thread(s)
#> _________________________________________________________________________
#> _________________________________________________________________________
#> ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.Using the “run_multiple_get_results” function, extract
the results from the model object that was simulated 50 times for
comparison across simulations.
# Retrieving the results
ans <- run_multiple_get_results(model_seirconn, nthreads = 2)
head(ans$total_hist)
#> sim_num date nviruses state counts
#> 1 1 0 1 Susceptible 9999
#> 2 1 0 1 Exposed 1
#> 3 1 0 1 Infected 0
#> 4 1 0 1 Recovered 0
#> 5 1 1 1 Susceptible 9999
#> 6 1 1 1 Exposed 1
head(ans$reproductive)
#> sim_num virus_id virus source source_exposure_date rt
#> 1 1 0 COVID-19 9700 0 0
#> 2 1 0 COVID-19 -1 0 1
#> 3 2 0 COVID-19 9964 50 0
#> 4 2 0 COVID-19 9828 50 0
#> 5 2 0 COVID-19 9689 50 0
#> 6 2 0 COVID-19 9687 50 0Plotting
To plot the epicurves and reproductive numbers over time using
boxplots, extract the results from the model object using
“ans”. For this example, the dates are filtered to be less
than or equal to 20 to observe the epicurves in the first 20 days.
Notice each boxplot in the below table represents the observed values
from each of the 50 simulations for each date.
seirconn_50 <- ans$total_hist
seirconn_50 <- seirconn_50[seirconn_50$date <= 20, ]
plot(seirconn_50)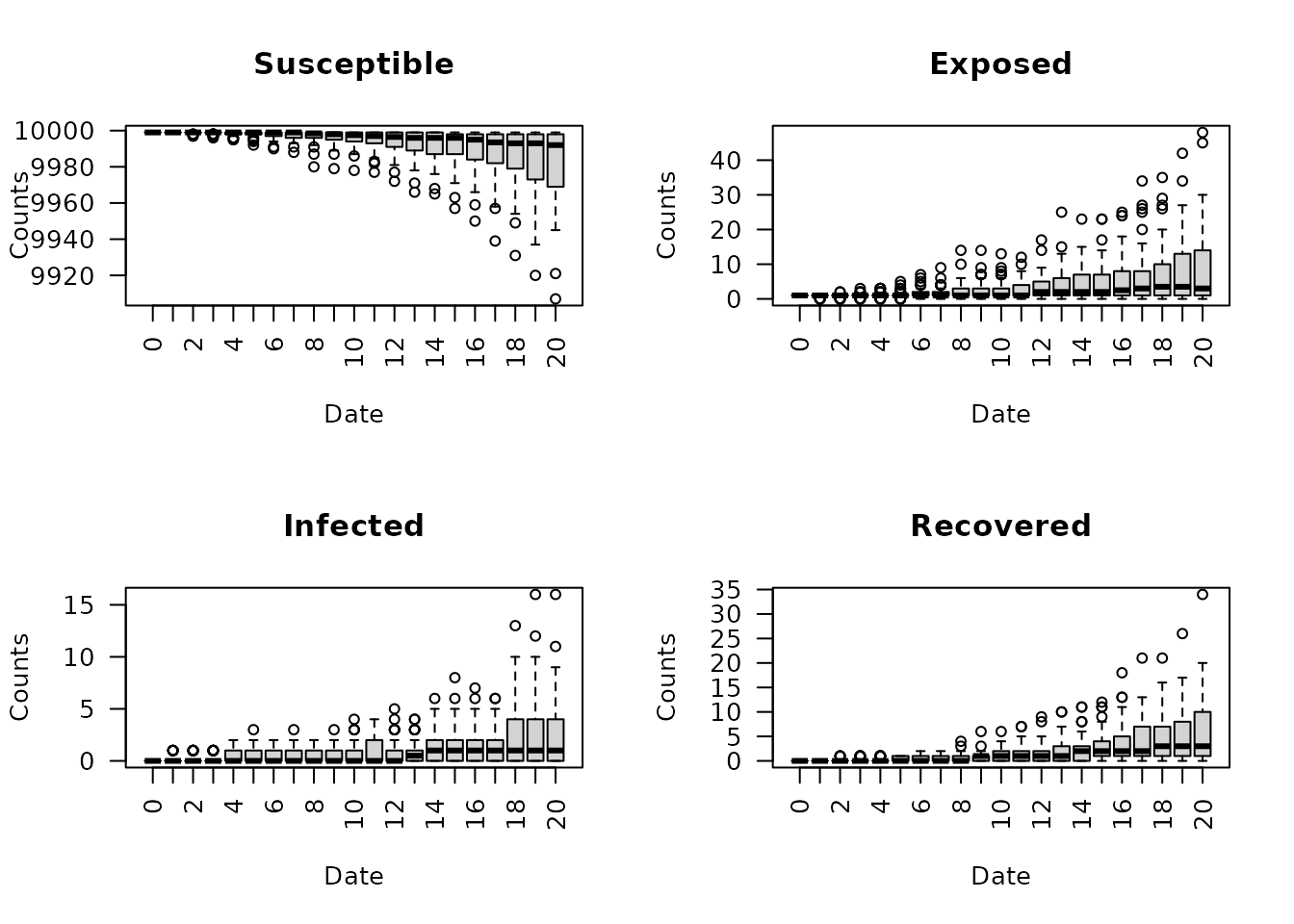
To view the a plot of the reproductive number over all 50 days for
each of the 50 simulations, store the reproductive results to a new
object using “ans”, then plot using the “boxplot” function.
Notice each source exposure date displays a boxplot representing the
distribution of reproductive numbers across all 50 simulations. As
expected, the reproductive number on average, decreases over time.
seirconn_50_r <- ans$reproductive
plot(seirconn_50_r)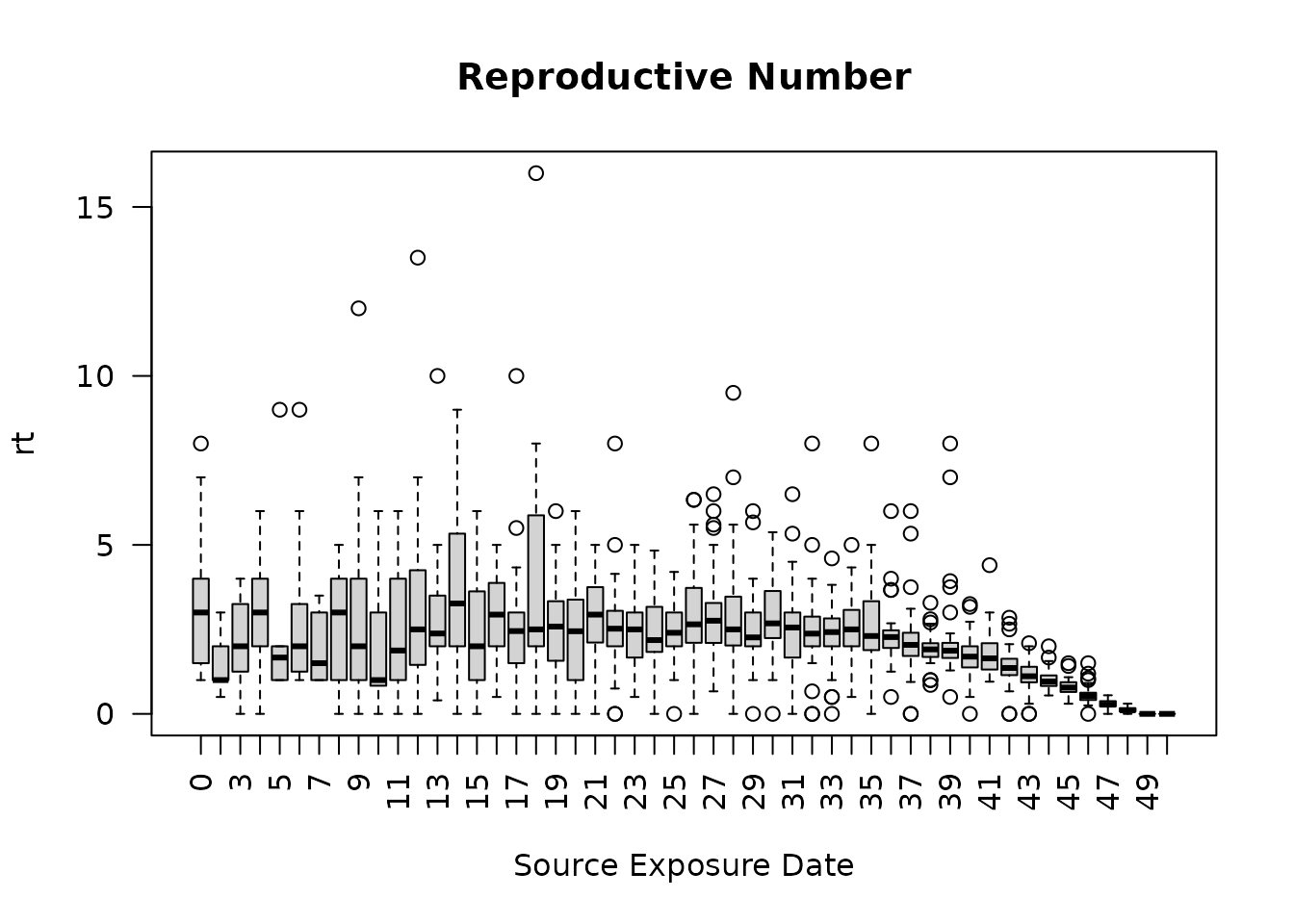
# boxplot(rt ~ source_exposure_date, data = seirconn_50_r,
# main = "Reproductive Number",
# xlab = "Source Exposure Date",
# ylab = "rt",
# border = "black",
# las = 2)