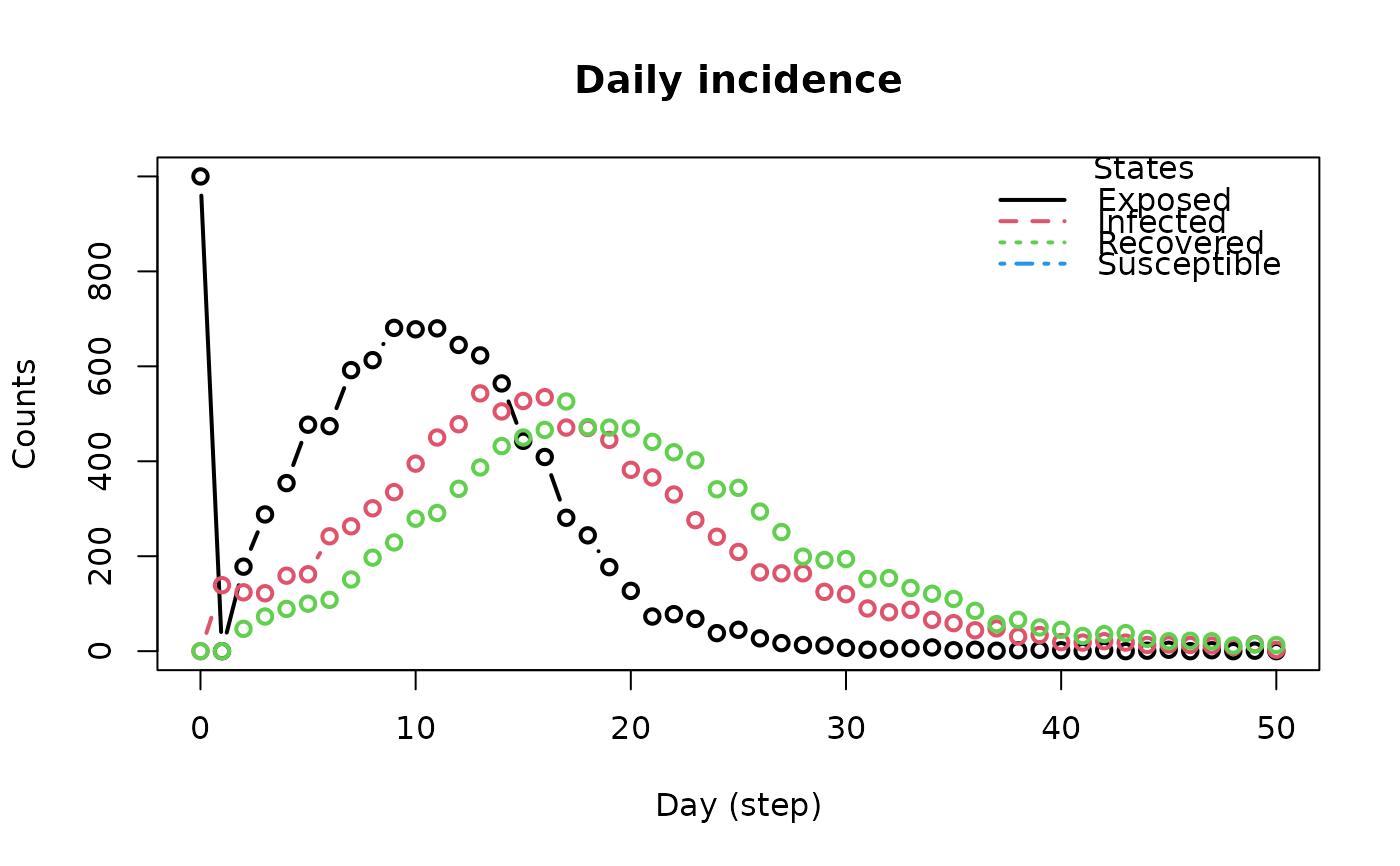
Functions to extract and visualize state transition counts, daily incidence, and conversion to array format.
Arguments
- x
An object of class
epiworld_sir,epiworld_seir, etc. (any model), or an object of class epiworld_hist_transition.- skip_zeros
Logical scalar. When
FALSEit will return all the entries in the transition matrix.- ...
In the case of plot methods, further arguments passed to graphics::plot.
- ylab, xlab, main, type
Further parameters passed to
graphics::plot()- plot
Logical scalar. If
TRUE(default), the function will plot the desired statistic.
Value
get_hist_transition_matrixreturns a data.frame with four columns: "state_from", "state_to", "date", and "counts."
The
as.arraymethod forepiworld_hist_transitionobjects turns thedata.framereturned byget_hist_transition_matrixinto an array ofnstates x nstates x (ndays + 1)entries, where the first entry is the initial state.
The
plot_incidencefunction returns a plot originating from the objectget_hist_transition_matrix.
The
plotmethod forepiworld_hist_transitionreturns a plot of the daily incidence.
Details
The plot_incidence function is a wrapper between
get_hist_transition_matrix and its plot method.
The plot method for the epiworld_hist_transition class plots the
daily incidence of each state. The function returns the data frame used for
plotting.
Examples
# SEIR Connected model
seirconn <- ModelSEIRCONN(
name = "Disease",
n = 10000,
prevalence = 0.1,
contact_rate = 2.0,
transmission_rate = 0.8,
incubation_days = 7.0,
recovery_rate = 0.3
)
set.seed(937)
run(seirconn, 50)
#> _________________________________________________________________________
#> |Running the model...
#> |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
#> |
# Get the transition history
t_hist <- get_hist_transition_matrix(seirconn)
head(t_hist)
#> state_from state_to date counts
#> 1 Susceptible Susceptible 0 9000
#> 2 Exposed Susceptible 0 0
#> 3 Infected Susceptible 0 0
#> 4 Recovered Susceptible 0 0
#> 5 Susceptible Exposed 0 1000
#> 6 Exposed Exposed 0 0
# Convert to array
as.array(t_hist)[, , 1:3]
#> , , 0
#>
#> Susceptible Exposed Infected Recovered
#> Susceptible 9000 1000 0 0
#> Exposed 0 0 0 0
#> Infected 0 0 0 0
#> Recovered 0 0 0 0
#>
#> , , 1
#>
#> Susceptible Exposed Infected Recovered
#> Susceptible 9000 0 0 0
#> Exposed 0 861 139 0
#> Infected 0 0 0 0
#> Recovered 0 0 0 0
#>
#> , , 2
#>
#> Susceptible Exposed Infected Recovered
#> Susceptible 8822 178 0 0
#> Exposed 0 737 124 0
#> Infected 0 0 92 47
#> Recovered 0 0 0 0
#>
# Plot incidence
inci <- plot_incidence(seirconn)