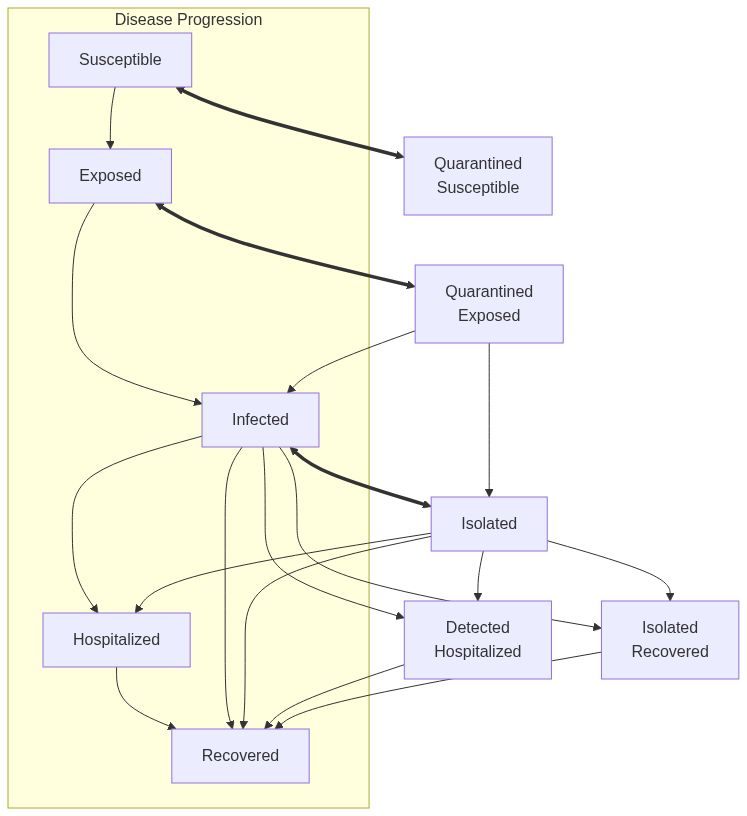
Susceptible Exposed Infected Removed model (SEIR) with mixing and quarantine
Source:R/ModelSEIRMixingQuarantine.R
ModelSEIRMixingQuarantine.RdModelSEIRMixingQuarantine creates a model of the SEIR type with mixing and
a quarantine mechanism. Agents who are infected can be quarantined or
isolated. Isolation happens after the agent has been detected as infected,
and agents who have been in contact with the detected person will me moved
to quarantined status.
Usage
ModelSEIRMixingQuarantine(
name,
n,
prevalence,
contact_rate,
transmission_rate,
incubation_days,
recovery_rate,
contact_matrix,
hospitalization_rate,
hospitalization_period,
days_undetected,
quarantine_period,
quarantine_willingness,
isolation_willingness,
isolation_period,
contact_tracing_success_rate,
contact_tracing_days_prior
)Arguments
- name
String. Name of the virus
- n
Number of individuals in the population.
- prevalence
Double. Initial proportion of individuals with the virus.
- contact_rate
Numeric scalar. Average number of contacts per step.
- transmission_rate
Numeric scalar between 0 and 1. Probability of transmission.
- incubation_days
Numeric scalar. Average number of days in the incubation period.
- recovery_rate
Numeric scalar between 0 and 1. Probability of recovery.
- contact_matrix
Matrix of contact rates between individuals.
- hospitalization_rate
Double. Rate of hospitalization.
- hospitalization_period
Double. Period of hospitalization.
- days_undetected
Double. Number of days an infection goes undetected.
- quarantine_period
Integer. Number of days for quarantine.
- quarantine_willingness
Double. Proportion of agents willing to quarantine.
- isolation_willingness
Double. Proportion of agents willing to isolate.
- isolation_period
Integer. Number of days for isolation.
- contact_tracing_success_rate
Double. Probability of successful contact tracing.
- contact_tracing_days_prior
Integer. Number of days prior to the onset of the infection for which contact tracing is effective.
Value
The
ModelSEIRMixingQuarantinefunction returns a model of class epiworld_model.
Details
The contact_matrix is a matrix of contact rates between entities. The
matrix should be of size n x n, where n is the number of entities.
This is a row-stochastic matrix, i.e., the sum of each row should be 1.
The quarantine and isolation processes can be turned off by specifying
quarantine_period < 0 and isolation_period < 0 respectively. In other
words, any negative value for these parameters will suppress the
corresponding process.
The initial_states function allows the user to set the initial state of the model. In particular, the user can specify how many of the non-infected agents have been removed at the beginning of the simulation.
See also
epiworld-methods
Other Models:
ModelDiffNet(),
ModelSEIR(),
ModelSEIRCONN(),
ModelSEIRD(),
ModelSEIRDCONN(),
ModelSEIRMixing(),
ModelSIR(),
ModelSIRCONN(),
ModelSIRD(),
ModelSIRDCONN(),
ModelSIRLogit(),
ModelSIRMixing(),
ModelSIS(),
ModelSISD(),
ModelSURV()
Examples
# Start off creating three entities.
# Individuals will be distributed randomly between the three.
e1 <- entity("Population 1", 3e3, as_proportion = FALSE)
e2 <- entity("Population 2", 3e3, as_proportion = FALSE)
e3 <- entity("Population 3", 3e3, as_proportion = FALSE)
# Row-stochastic matrix (rowsums 1)
cmatrix <- c(
c(0.9, 0.05, 0.05),
c(0.1, 0.8, 0.1),
c(0.1, 0.2, 0.7)
) |> matrix(byrow = TRUE, nrow = 3)
N <- 9e3
flu_model <- ModelSEIRMixingQuarantine(
name = "Flu",
n = N,
prevalence = 1 / N,
contact_rate = 20,
transmission_rate = 0.1,
recovery_rate = 1 / 7,
incubation_days = 7,
contact_matrix = cmatrix,
hospitalization_rate = 0.05,
hospitalization_period = 7,
days_undetected = 3,
quarantine_period = 14,
quarantine_willingness = 0.8,
isolation_period = 7,
isolation_willingness = 0.5,
contact_tracing_success_rate = 0.7,
contact_tracing_days_prior = 3
)
# Adding the entities to the model
flu_model |>
add_entity(e1) |>
add_entity(e2) |>
add_entity(e3)
set.seed(331)
run(flu_model, ndays = 100)
#> _________________________________________________________________________
#> Running the model...
#> ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||| done.
summary(flu_model)
#> ________________________________________________________________________________
#> ________________________________________________________________________________
#> SIMULATION STUDY
#>
#> Name of the model : SEIR with Mixing and Quarantine
#> Population size : 9000
#> Agents' data : (none)
#> Number of entities : 3
#> Days (duration) : 100 (of 100)
#> Number of viruses : 1
#> Last run elapsed t : 21.00ms
#> Last run speed : 41.68 million agents x day / second
#> Rewiring : off
#>
#> Global events:
#> - Update infected individuals (runs daily)
#>
#> Virus(es):
#> - Flu
#>
#> Tool(s):
#> (none)
#>
#> Model parameters:
#> - Avg. Incubation days : 7.0000
#> - Contact rate : 20.0000
#> - Contact tracing days prior : 3.0000
#> - Contact tracing success rate : 0.7000
#> - Days undetected : 3.0000
#> - Hospitalization period : 7.0000
#> - Hospitalization rate : 0.0500
#> - Isolation period : 7.0000
#> - Isolation willingness : 0.5000
#> - Prob. Recovery : 0.1429
#> - Prob. Transmission : 0.1000
#> - Quarantine period : 14.0000
#> - Quarantine willingness : 0.8000
#>
#> Distribution of the population at time 100:
#> - ( 0) Susceptible : 8999 -> 8998
#> - ( 1) Exposed : 1 -> 0
#> - ( 2) Infected : 0 -> 0
#> - ( 3) Isolated : 0 -> 0
#> - ( 4) Detected Hospitalized : 0 -> 0
#> - ( 5) Quarantined Susceptible : 0 -> 0
#> - ( 6) Quarantined Exposed : 0 -> 0
#> - ( 7) Isolated Recovered : 0 -> 0
#> - ( 8) Hospitalized : 0 -> 0
#> - ( 9) Recovered : 0 -> 2
#>
#> Transition Probabilities:
#> - Susceptible 1.00 0.00 - - - 0.00 - - - -
#> - Exposed - 0.82 0.18 - - - - - - -
#> - Infected - - - 0.50 - - - - - 0.50
#> - Isolated - - - 0.67 - - - 0.33 - -
#> - Detected Hospitalized - - - - - - - - - -
#> - Quarantined Susceptible 0.07 - - - - 0.93 - - - -
#> - Quarantined Exposed - - - - - - - - - -
#> - Isolated Recovered - - - - - - - 0.67 - 0.33
#> - Hospitalized - - - - - - - - - -
#> - Recovered - - - - - - - - - 1.00
#>